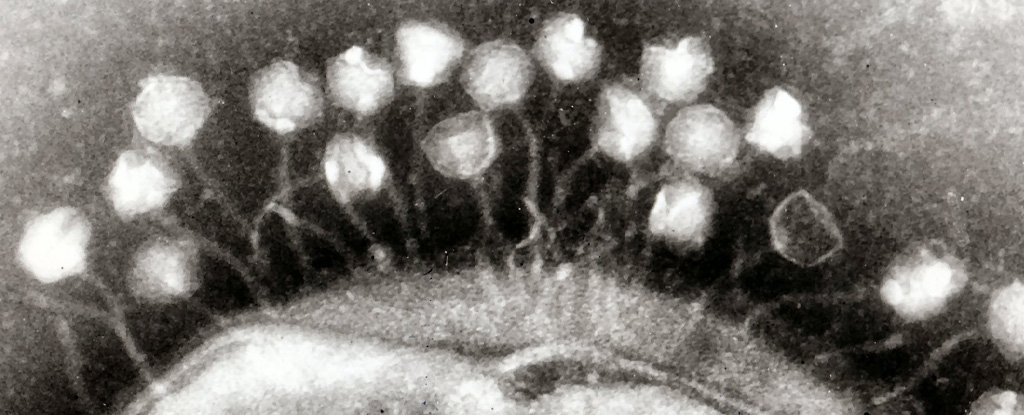
Research published today in Natural microbiology identified 54,118 virus species living in the human gut – 92 percent of which were previously unknown.
But as we and our colleagues at the Joint Genome Institute and Stanford University in California discovered, the vast majority of them were bacteriophages, or “phages” for short. These viruses “eat†bacteria and cannot attack human cells.
When most of us think of viruses, we think of the organisms that infect our cells with diseases such as mumps, measles or, more recently, COVID-19. However, there are a large number of these microscopic parasites in our bodies – mainly in our gut – which target the microbes that live there.
Everyone poops (but not all poop is the same)
There has recently been a lot of interest in the human gut microbiome: the collection of microorganisms that live in our gut.
In addition to helping us digest our food, these microbes have many other important roles. They protect us against pathogenic bacteria, modulate our mental well-being, prime our immune system when we are children, and play an ongoing role in immune regulation in adulthood.
It’s fair to say that the human gut is now the best-studied microbial ecosystem on the planet. Yet more than 70 percent of the microbial species that live there have not yet been cultured in the laboratory.
We know this because we can access the genetic blueprints of the gut microbiome through an approach known as metagenomics. It is a powerful technique whereby DNA is directly extracted from an environment and randomly sequenced, giving us insight into what is inside and what it might do.
Metagenomic studies have revealed how far we still have to go to catalog and isolate all microbial species in the human gut – and even further when it comes to viruses.
11,810 poop samples
In our new research, we and our colleagues computer-extracted viral sequences from 11,810 publicly available fecal metagenomas collected from people in 24 different countries. We wanted to get a sense of the extent to which viruses have taken up residence in the human gut.
This effort resulted in the Metagenomic Gut Virus Catalog, the largest such resource to date. This catalog includes 189,680 viral genomes which represent more than 50,000 distinct viral species.
Remarkably (but perhaps predictably), over 90 percent of these viral species are new to science. They collectively encode more than 450,000 distinct proteins – a huge reservoir of functional potential that can be beneficial or harmful to their microbial hosts, and in turn humans.
We also delved into the subspecies of different viruses and found that some exhibited striking geographic patterns in the 24 countries studied.
For example, a subspecies of the recently described and enigmatic crAssphage was widespread in Asia, but was rare or absent in samples from Europe and North America. This may be due to the localized expansion of this virus in specific human populations.
One of the most common functions we discovered on our molecular field trip were diversity-generating retroelements (DGRs). This is a class of genetic elements that mutate specific target genes to generate variation that may be beneficial to the host. In the case of RDGs in viruses, this may aid in the ongoing evolutionary arms race with their bacterial hosts.
Surprisingly, we have found that a third of the most common viral proteins have unknown functions, including more than 11,000 genes far removed from “beta-lactamases”, which allow resistance to antibiotics such as penicillin.
Linking intestinal viruses to their microbial hosts
After identifying the phages, our next task was to link them to their microbial hosts. CRISPRs, best known for their many applications in gene editing, are bacterial immune systems that “remember†past viral infections and prevent them from reproducing.
They do this by copying and storing fragments of the invading virus in their own genomes, which can then be used to specifically target and destroy the virus in future encounters.
We have used this record of past attacks to link numerous viral sequences to their hosts in the intestinal ecosystem. Not surprisingly, very abundant viral species were linked to very abundant bacterial species in the gut, mainly belonging to the bacterial phyla Firmicutes and Bacteroidota.
So what can we do with all of this new information? A promising application of an inventory of intestinal viruses and their hosts is phage therapy. Phage therapy is an ancient concept prior to antibiotics, in which viruses are used to selectively target bacterial pathogens to treat infections.
There has been talk of the possibility of personalizing people’s gut microbiomes with the help of dietary interventions, probiotics, prebiotics or even “transpoosions” (faecal microbiota transplants), to improve the health of an individual.
Phage therapy can be a useful adjunct to this goal, adding species or even subspecies level precision to microbiome manipulation. For example, the bacterial pathogen Clostridioides difficile (or Cdiff for short) is a major cause of nosocomial diarrhea that could be specifically targeted by phages.
More subtle manipulation of non-pathogenic bacterial populations in the gut may be achievable with phage therapy. A complete collection of intestinal viruses is a useful first step for such applied purposes.
It should be noted, however, that our data projections suggest that we have only studied a fraction of the total gut viral diversity. So we still have a long way to go.
Philip Hugenholtz, Professor of Microbiology, School of Chemistry and Molecular Biosciences, University of Queensland and Soo Jen Low, Postdoctoral Fellow, University of Queensland.
This article is republished from The Conversation under a Creative Commons license. Read the original article.